Christina Woo works in her Lab in the Conant Building.
Kris Snibbe/Harvard file photo
Dissecting the ‘undruggable’
Researchers design new highly-selective tool to study proteins through sugars they depend on
Sugar has been called “evil,” “toxic,” and “poison.” But the body needs sugars, too. Sugar molecules help cells recognize and fight viruses and bacteria, shuttle proteins from cell to cell, and make sure those proteins function. Too much or too little can contribute to a range of maladies, including neurodegenerative diseases like Alzheimer’s, inflammation, diabetes, and even cancer.
About 85 percent of proteins, including those associated with Alzheimer’s and Parkinson’s, are beyond the reach of current drugs. One critical and abundant sugar (O-GlcNAc, pronounced o-glick-nack) is found on more than 5,000 proteins, many of them considered “undruggable.” But now, researchers at Harvard have designed a new, highly selectiveO-GlcNAc pencil and eraser — tools that can add or remove the sugar from a protein with no off-target effects — to examine exactly what these sugars are doing and, eventually, engineer them into new treatments for the undruggable.
“We can now start studying particular proteins and see what happens when you add or remove the sugar,” said Daniel Ramirez, a co-author on the paper published in Nature Chemical Biology and a Ph.D. candidate in biological and biomedical sciences in the Graduate School of Arts and Sciences. “This is turning out to be very important for a lot of chronic diseases, like cancer and diabetes and Alzheimer’s.”
Ramirez designed the original O-GlcNAc pencil, which was reported in ACS Chemical Biology.
All cells carry a multitude of sugars (called glycans), but they’re notoriously hard to study. Current tools provide either a wide-lens view (turning on or off all the O-GlcNAcs in a cell) or an ultra-zoomed-in view (turning on or off a single sugar on one amino acid on one protein). Neither of these perspectives can show the crucial insight that would enable researchers to connect the dots from O-GlcNAc to disease: what O-GlcNAc molecules are doing to a protein as a whole.
“With the protein-level approach, we’re filling in an important piece that was missing,” said Christina Woo, an associate professor of chemistry and chemical biology, who led the study. Her lab’s tool is like Goldilocks’ lukewarm bowl of porridge: Not too broad, not too specific. Just right.
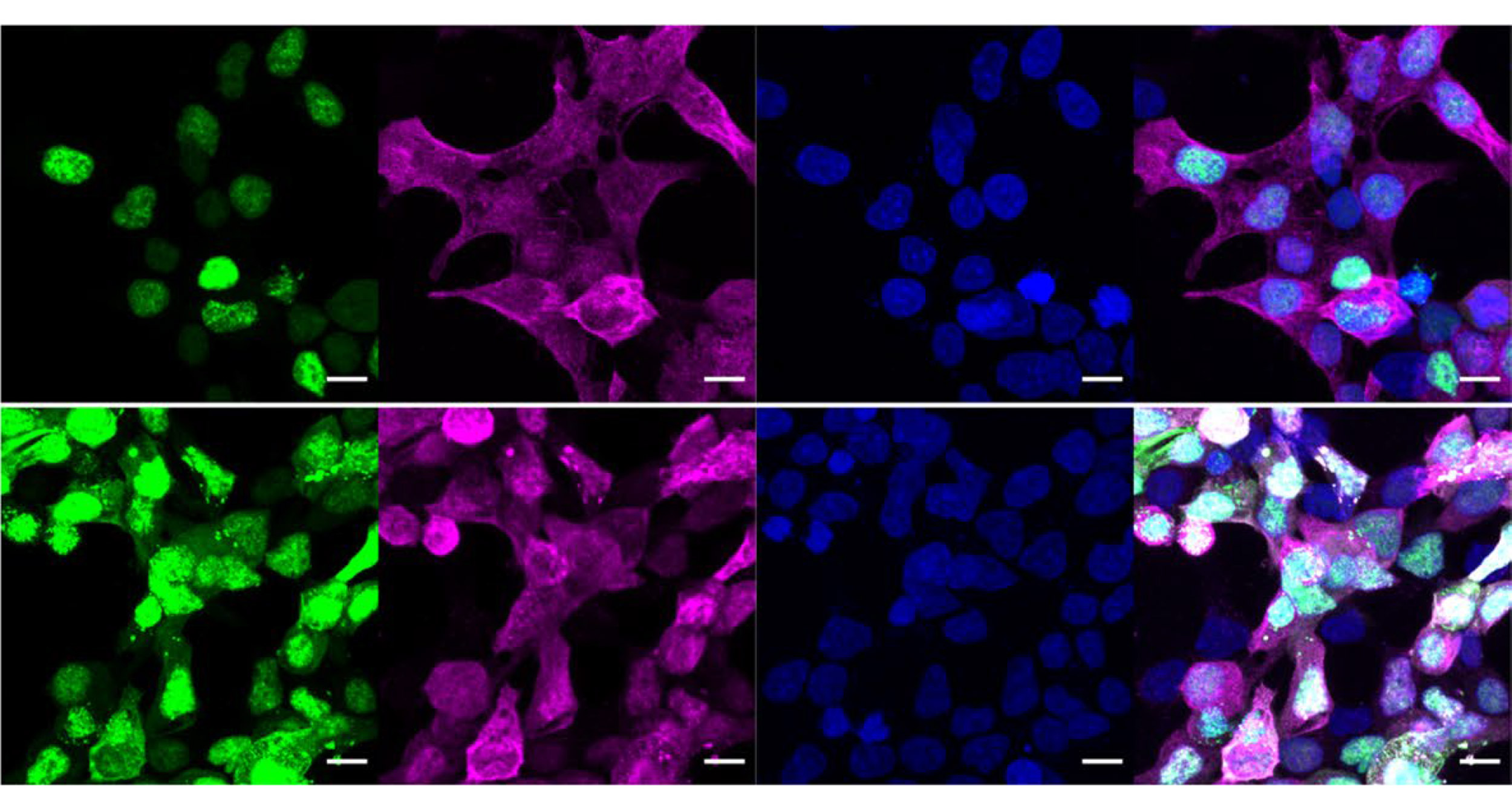
Fluorescent imaging of proteins helped the research team determine how effective their O-GlcNAc pencil and eraser tools were at writing and removing sugars from their protein targets and what changes occurred to the protein after their alteration.
Image courtesy of the Woo Lab
“Once you have any protein of interest, you can apply this tool on that protein and look at the outcomes directly,” said first author and postdoctoral scholar Yun Ge. Ge engineered the O-GlcNAc eraser, which, like the pencil, uses a nanobody as a protein homing device. The tool is adaptable, too; As long as a nanobody exists for a protein of choice, the tool can be modified to target any protein for which there is a homing nanobody.
The nanobody is a crucial component, but it has limitations: Whether it remains stuck to the target protein is still in question, and the molecule could alter the function or structure of the protein once stuck. If cellular changes can’t be definitively linked to the sugar on the protein, that muddies the data.
To skirt these potential limitations, the team engineered O-GlcNAc pencils and erasers to be “catalytically dead,” said Woo. The neutered enzymes won’t make unwanted changes along the way to their target protein. And they can both add and remove sugars, unlike previous tools, which cause permanent changes. Of course, once they connect a specific protein function to O-GlcNAc, they can then use those tools to zoom in and locate exactly where those sugars are latching onto and modifying the protein.
Already, a few of the Woo lab’s collaborators are using the pencil/eraser combo to study O-GlcNAc in live animals. One, for example, is using fruit flies to study how the sugar impacts a protein associated with Alzheimer’s disease. The sugar is also associated with Parkinson’s disease progression: “If you’re taking in less glucose, then you’re not able to produce this sugar inside the cells,” said Ramirez. “That means the body can’t attach the sugars to the proteins, which causes wide-reaching changes to the cells, aggravating the disease. In diabetes, excess sugars cause similar global disruption, and cancer cells tend to eat lots of sugars. Now, with the Woo lab’s pencil/eraser pair, researchers can identify exactly how these sugars impact various proteins and start to design drugs to reverse negative effects.
Next, the team plans to tweak the tool to achieve even greater control. With optogenetics, for example, they could switch sugars on or off with just a flash of light. Swapping out nanobodies for small molecules (used in traditional drug design), they could edge closer to new treatments. They’re also designing an eraser for the eraser — a tool with a kill switch — and plan to incorporate nanobodies that can target a naturally occurring protein (for this study, they tagged proteins so the nanobody could find them). “We’re basically trying to make the system more natural and function the way the cell does,” said Ramirez.
Woo also plans to investigate how O-GlcNAc may influence traditionally undruggable proteins called transcription factors, which turn genes on and off. If O-GlcNAc plays a role in that process, the sugars could be engineered to study and regulate gene function, too.
“We really don’t know what people are going to find once we give them these tools,” said Ramirez. The tool may be new, but the potential is great: “We’re on the first-generation iPhone, basically,” he continued, “but we’re already working on the next couple generations.”




